Fall Meet-and-Greet!
Students, faculty, and staff gathered on the first Friday of fall classes to say hello, meet first-year and transfer students, and eat pizza and cookies. And now for a wonderful fall semester!
Bard Biology Club visits the Cary Institute of Ecosystem Studies
Bard Biology Club members visited the Cary Institute of Ecosystem Studies in Millbrook, New York in April. The trip, organized by club members, gave students the opportunity to talk with scientists, educators, and science writers about careers in science. The students also toured the grounds and saw the Institute's newly-renovated facilities dedicated to the study of ecology and the environment.
Professor Todd publishes paper on emerging fungal pathogen
|
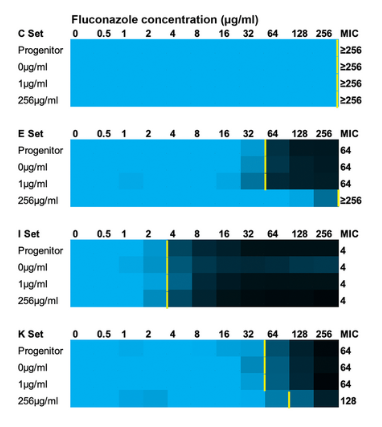
Professor Rob Todd and his colleagues published a paper that explores anti-fungal drug resistance, a critical threat to global public health. In the newly emerging fungal pathogen, Candida auris, anti-fungal drug resistance to all three major classes of anti-fungal drugs is on the rise. To better understand how drug resistance arises within this pathogen, Todd and his colleagues conducted an in vitro evolution experiment in which 17 different human clinical isolates were exposed to varying concentrations of drug. They identified multiple different pathways in which this organism undergoes genome modifications (mutations, increases of chromosome copy number, and chromosome size alterations) that were associated with decreases in sensitivity to the most commonly prescribed anti-fungal drug, Fluconazole. Some of these genome modifications occurred in as few as 30 generations (3 days). They also identified one human clinical isolate that exhibited an increased mutation rate when compared to the other clinical isolates, and that isolate had significantly decreased sensitivity to Fluconazole. Together, these findings indicate that initial genetic diversity can have major impacts on the evolution of drug resistance in this critically important emerging pathogen. Find the full paper here.
|
"Mammals" class documents campus wildlife
Students in the fall class on "Mammals", taught by Professor Keesing, placed wildlife cameras around campus and documented this handsome coyote, photographed sniffing a can of cat food that worked perfectly as bait for the camera. In addition to capturing lots of pastoral images of deer and squirrels, the students also took great photos of a raccoon, some red foxes, and even an elusive gray fox.
New paper on bacterial communities in wastewater discharge
|
Associate Professor of Biology Gabriel G. Perron, and Bard biology graduates Daniella Azulai ’17 and Mary Reid ’21 have co-published a new study, “Bacteria communities and water quality parameters in riverine water and sediments near wastewater discharges,” in the journal Scientific Data. Over five months, the team monitored microbial contaminants relating to the treated water outflow of the wastewater treatment plant operated by Bard, which releases into the Saw Kill, a tributary of the Hudson River and also the source of freshwater for the campus. This is the first of many datasets and research papers that they hope to publish on Bard’s water system. Preliminary analyses provide insight into the impacts of watershed-wide usage of the Saw Kill as both drinking water source and treated sewage receiver. Future use of this dataset will include a focus on antibiotic resistant bacterial genes.
|
Keesing receives International Cosmos Prize
|
Professor of Biology Felicia Keesing has been selected as the winner of the International Cosmos Prize for 2022. Dr. Keesing will receive a certificate of merit, a medallion, and a monetary prize of 40 million yen at the award ceremony, which will be held in Osaka, Japan, on November 9. Dr. Keesing will also give a commemorative lecture, participate in a symposium held in her honor, and have an audience with the Emperor and Empress of Japan.
The annual Cosmos Prize is awarded in recognition of a body of work that has significantly advanced our understanding of the relationships among living organisms and the interdependence of life and the global environment. The decision to award the prize to Dr. Keesing was reached after the committee evaluated 174 nominations from 28 countries. Previous recipients include Jared Diamond (1998), David Attenborough (2000), E.O. Wilson (2012), and Jane Goodall (2017). |
Professor Kaishian urges fungal conservation
In a new paper in The Conversation, Professor Patty Kaishian, Visiting Assistant Professor of Biology, and her colleagues argue that the conservation of fungi is critical. Fungi, they argue, are poorly studied -- scientists have identified only ~150,000 of an estimated 2-4 million fungal species. And yet we know that fungi play crucial roles in ecosystems, functions that humans have put to use in the making of many of our favorite foods, including beer, wine, and soy sauce, as well as the manufacture of important medicines like penicillin. Mycologists -- scientists who study fungi -- want these important creatures included in biodiversity targets to be set later this year in China.
Alumni return for grad panel
Three recent Bard biology alumni joined us for a panel discussion on careers. Sacha Medjo-Akono '21 works as an educator at the New York Aquarium. Quanita Kendrick '17 works for the US Army Corps of Engineers on environmental justice initiatives. Biz Osborne-Schwartz is an outdoor educator and teacher in the western US, leading trips for students of all ages. The three gave students advice about particularly useful experiences they had at Bard -- including teaching, tutoring, and working with groups of people outside of the classroom (such as through being a peer counselor). They encouraged students to keep trying new things, and to be open to the possibilities in front of them, because each new experience can have unexpected benefits later on.
Plant Ecology: Bard students install pollinator gardens with 2nd graders
Students in Professor Cathy Collins' Plant Ecology course installed pollinator gardens with 2nd graders from Edson Elementary school in Kingston. Bard students researched native plant species that are pollinated by different species and that flower at different times of year. From this list, each class at Edson selected 6-10 plant species based on which pollinators they wanted to attract. Bard students visited Edson two times. The first time they played a game to teach kids about traits of flowers that attract certain pollinators. The second visit they taught the kids about plant life cycles and sowed the seeds into the garden. The 2nd grade teachers assigned work to their students related to the project, and with the school librarian, students pay regular visits to the gardens to collect data!
Alumni panel on graduate school
Four Bard biology alumni joined current Bard biology students for a panel discussion on graduate school options. Kelsey O'Brien '17 is a graduate student in cell and molecular biology at the University of Pennsylvania Medical School. Liz Miller '17 is a Ph.D. student in oceanography at the University of Hawaii. Adenike Akapo '13 is a third-year dental student at New York University. Lizzy Elliot '18 is a first-year Master's student in ecology at the College of William and Mary. The four gave students advice about choosing programs of study, applying to graduate school, finding funding, and staying open to new possibilities, including unexpected opportunities in unexpected places. Common themes for current students included valuing the skills they learned about writing and giving presentations, and recognizing that you don't have to know immediately what you want to do to be successful.
A new paper from the Keesing Lab
|
A new review paper from Felicia Keesing explores how diversity can affect pathogen transmission. For many diseases of plants and animals, including humans, the presence of low-quality hosts reduces the overall transmission of disease-causing pathogens. People have used these “dilution effects” to manage diseases for over a century. Recent evidence demonstrates that dilution effects also occur naturally, protecting us from greater risk of being exposed to infectious diseases. When biodiversity declines, these natural dilution effects disappear, providing a powerful link between the conservation of biodiversity and the health of humans, wildlife, and plants. Publication info: Keesing, F., & Ostfeld, R. S. (2021). Dilution effects in disease ecology. Ecology Letters. PDF
|
Professor Dueker on citizen science and environmental justice
|
Citizen science has seen a boon in participation by people motivated by the loss of environmental protections and the impacts of Covid-19 on data collection and justice.
“There isn’t enough environmental monitoring to begin with, and it will only decrease,” said Eli Dueker, a professor of environmental and urban studies at Bard College. “So what we end up with is community scientists often working with research scientists to fill that gap. And that can be really effective because it allows communities to know the pollution hot spots with both air and water.” FULL TEXT |
Devin Fraleigh '18, Jackson Barratt Heitmann '18, and Bruce Robertson publish paper on evolutionary traps
Biology alums Devin Fraleigh '18 and Jackson Barratt Heitmann '18 have joined Professor Bruce Robertson to publish the results of their research in the journal Animal Behavior.
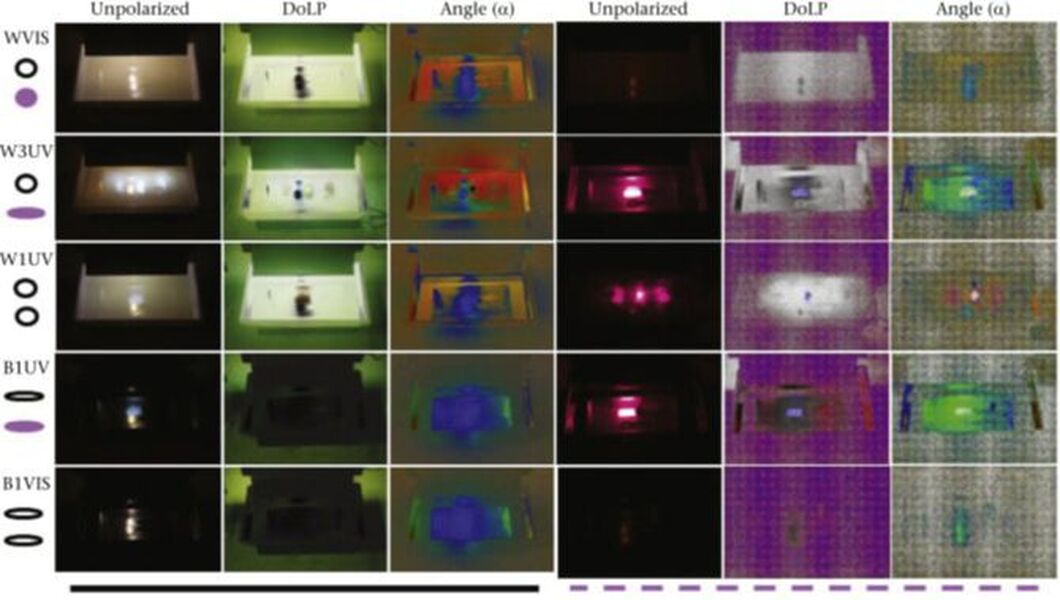
“When animals are misguided by evolved behavioral cues to preferentially make mistakes, they are caught in an evolutionary trap,” write Associate Professor of Biology Bruce Robertson, Devin C. Fraleigh ’18, and Jackson Barratt Heitmann ’18 in a newly published scientific paper. “Aquatic insects rely heavily on polarized light cues to locate bodies of water necessary for laying eggs and mating. However, where artificial objects (e.g. asphalt, buildings) are at least as effective at polarizing light as natural water bodies, aquatic insects may instead prefer to lay eggs on those surfaces where their eggs fail to hatch.”
Published in Animal Behavior, their paper surveyed natural and artificial environment to understand the properties of objects that can polarize natural and artificial sources of UV light. They conducted a field experiment to test the importance of UV polarized light in guiding habitat selection behavior in six families of aquatic insects. The results highlight a quantitatively new type of ecological light pollution capable of creating evolutionary traps for insects at night, or even during the day.
Read the full study here: Fraleigh, Devin C., Jackson, Barratt Heitmann, and Bruce A. Robertson. "Ultraviolet polarized light pollution and evolutionary traps for aquatic insects." Animal Behaviour (2021).
“When animals are misguided by evolved behavioral cues to preferentially make mistakes, they are caught in an evolutionary trap,” write Associate Professor of Biology Bruce Robertson, Devin C. Fraleigh ’18, and Jackson Barratt Heitmann ’18 in a newly published scientific paper. “Aquatic insects rely heavily on polarized light cues to locate bodies of water necessary for laying eggs and mating. However, where artificial objects (e.g. asphalt, buildings) are at least as effective at polarizing light as natural water bodies, aquatic insects may instead prefer to lay eggs on those surfaces where their eggs fail to hatch.”
Published in Animal Behavior, their paper surveyed natural and artificial environment to understand the properties of objects that can polarize natural and artificial sources of UV light. They conducted a field experiment to test the importance of UV polarized light in guiding habitat selection behavior in six families of aquatic insects. The results highlight a quantitatively new type of ecological light pollution capable of creating evolutionary traps for insects at night, or even during the day.
Read the full study here: Fraleigh, Devin C., Jackson, Barratt Heitmann, and Bruce A. Robertson. "Ultraviolet polarized light pollution and evolutionary traps for aquatic insects." Animal Behaviour (2021).